Strengthening and microbial regulation mechanism of Bacillus on purification device for grass carp culture wastewater
- 1School of Civil Engineering and Transportation, South China University of Technology, Guangzhou, China
- 2Key Laboratory of Tropical and Subtropical Fishery Resource Application and Cultivation, Pearl River Fisheries Research Institute, Chinese Academy of Fishery Sciences, Guangzhou, China
- 3Guangdong Ecological Remediation of Aquaculture Pollution Research Center, Guangzhou, China
Aquaculture wastewater (AW) poses a threat to natural aquatic environments. Microecological agents are widely used to regulate and purify AW, with Bacillus being the most common. To evaluate the AW purification effect of adding Bacillus subtilis and Bacillus licheniformis to an AW treatment device, we constructed an experimental device including a small grass carp culture pond and three groups of cuboid reactors. The effects of adding the two strains to the AW treatment reactor on the AW purification effect and the microbiota compositions in the AW and packing surface biofilm were analyzed via high-throughput sequencing of the 16S rRNA gene. Our results showed that adding Bacillus bacteria to reactors improved the total nitrogen (TN) removal efficiency and reduced the chemical oxygen demand (COD). Adding both the B. subtillis and B. licheniformis preparations significantly increased the abundance of Firmicutes in the water microbiota of the reactor at the middle and end stages of the experiment. The addition of Bacillus changed the microbiota composition in the water and packing surface biofilm and significantly increased the abundance of Bacillus at the middle and later stages of the experiment. Therefore, the addition of Bacillus improved the TN removal efficiency in the AW grass carp treatment reactors and significantly reduced the COD in the AW by increasing the abundance of Bacillus and changing the microbiota composition in the system. We provide an effective way for improving the purification capacity of biofilm reactor.
Introduction
Aquatic products are rich in protein and various micronutrients, providing 15%–20% of the animal protein consumed by more than four billion people worldwide (Tezzo et al., 2021). Therefore, aquaculture plays an important role in our nutrition supply globally. China is a large producer of aquaculture (Wang et al., 2020). Chinese aquatic products have not only become an important part of China’s food supply but have also become an important source of global aquatic products. Currently, the main method of aquaculture in China is intensive aquaculture, which has the advantage of increasing the output of aquatic products and profits (Edwards, 2015). However, owing to the high stocking density of aquatic animal and large amount of feed input, the discharge of aquaculture wastewater (AW) causes eutrophication, hypoxia, and other adverse effects on the natural water environment (Li et al., 2019). Therefore, efficient treatment of pollutants in AW is necessary to ensure the sustainable development of the aquaculture industry.
The natural biofilm of aquatic environments refers to the surface film covering the sediment, stones, plant roots, and other appendages in the aquatic environment (Wu et al., 2011). It typically comprised algae, bacteria, fungi, protozoa, and epiphytes (Zippel et al., 2007) and is widely distributed in natural or artificial water environments such as rivers, lakes, wetlands, reservoirs, and ponds (Wu et al., 2019). Natural biofilms have a strong purification effect on water pollution (Ji et al., 2007; Wu et al., 2019). Currently, aquaculture farmers mostly add artificial fillers to filter ponds to stimulate the activity of indigenous microorganisms, increase the area of microbial attachment, and form a large number of biofilms to achieve the effect of purify AW (Wang et al., 2021). However, according to the current discharge of AW, natural biofilms in aquaculture ponds cannot completely purify AW (Li et al., 2019d). Owing to the limited purification effect of natural biofilms, enhancing their purification performance in aquaculture ponds is a promising research area. However, research on enhancing the purification performance of natural biofilms is scarce.
Bacillus spp. play an important role in water purification (Nayak, 2020). Bacillus subtilis is an aerobic heterotrophic bacterium with high digestive enzyme activity (Yi et al., 2020). Adding B. subtilis and Bacillus megaterium to the circulating water system can assist in reducing the ammonia nitrogen (NH4-N) content, chemical oxygen demand (COD), and fish mortality (Chen and Chen, 2001). Li et al. (2021) reported that B. subtilis could effectively reduce NH4-N and nitrite (NO2-N) contents in aquaculture water and had a certain removal effect on COD. Zhang (2011) reported that Bacillus could competitively inhibit the growth of dominant algae in water.
To optimize the microbiota composition of natural biofilms, strengthen the AW purification efficiency, and seek a simple, efficient, and practical method for AW purification, B. subtilis and Bacillus licheniformis were added to an AW purification device to explore the changes in the purification performance of AW and analyze the microbiota composition of the wastewater and biofilm in the packing surface of reactors. Our study will provide a reference for resolving pollution in the culture environment and suggest an effective way to improve the purification capacity of biofilm reactor, which will have great significance in green aquaculture in the future.
Materials and methods
Experimental devices and design
The experimental device included a small grass carp culture pond (2 × 2 × 1.5 m) and three groups of cuboid reactors, with an effective volume of approximately 61 L (50 × 35 × 35 cm) (Figure 1). There were 25 individuals grass carp in the culture pond. The initial average body weight of the grass carp was 0.35 ± 0.07 kg. The fish were fed with commercial feed (approximately 30% crude protein) at 9:00 and 16:00 each day. Fish were given approximately 1.5%–2.5% of their own weight in feed each time. The packing ratio of the three biofilm purification reactors was 55%. The packing in the reactors consisted of porous polyethylene (PPP; Figure 1) and porous spherical polypropylene (PSPP; Figure 1). The PSPP was a sphere with a diameter of 8 cm (Figure 1), and the PPP was a polyhedral column with a diameter of approximately 20 mm and a height of approximately 30 mm (Figure 1). Each PSPP sphere was packed using 20 PPP and each reactor was packed using 40 PSPP spheres, which were covered with a pressure net to fix the PSPP spheres at the bottom of the reactor. The wastewater from the aquaculture pond was pumped into the reactors through peristaltic pumps, with inlet and outlet-flow of 0.2 L/min. The theoretical hydraulic retention time was 5.4 h. The reactors were continuously exposed to air.
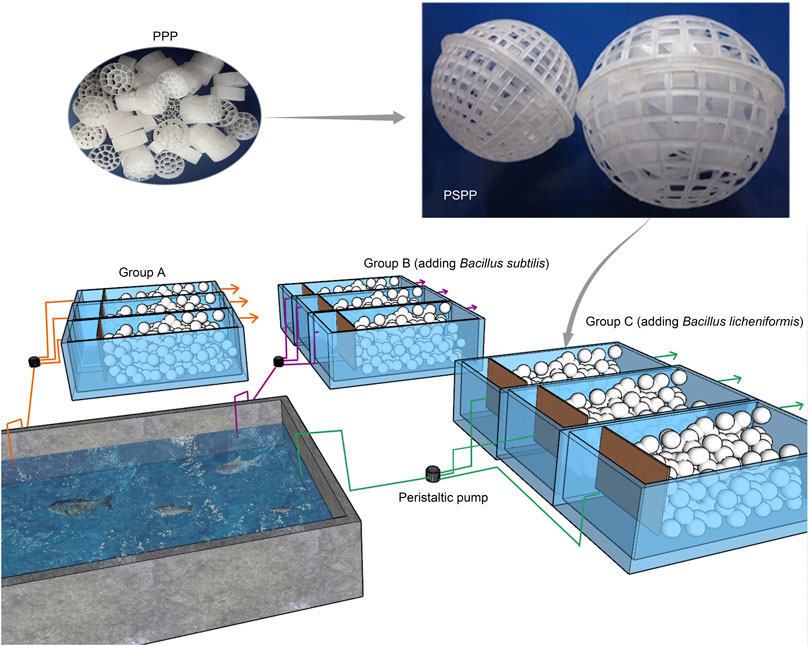
FIGURE 1. Diagram of experimental device. PPP, porous polyethylene packing; PSPP, porous spherical polypropylene packing.
The three groups of reactors were labeled A, B, and C, respectively. Each group consisted of three parallel reactors. The peristaltic pumps flowed water to form a natural biofilm on the surface of the packing, and the experiment was started. Group A was a natural biofilm group, and its biofilm was formed on the surface of the packing after wastewater from the aquaculture pond continuously entered the reactors. Groups B and C were enhanced biofilm groups. In addition to providing AW as in the group A reactors, 2.5 g of B. subtillis and B. licheniformis preparations was added to the two groups of devices every 3 days. The concentration of both microbial preparations was 3 × 109 cells/ g, and both were obtained from Bio-Form Ltd. (Guangdong, China). The experiment lasted for 42 days. The temperature and pH of the reactors were maintained at 25°C–32°C and 7.0–8.0 during operation. The water physicochemical parameters of the aquaculture pond and reactors at the beginning of the experiment were measured as initial values, and the water physicochemical parameters of the outlet of each reactor and pond were measured every 7 days. Water (W) and the biofilm of reactor packing (B) samples were collected at the early (E, 0 days after the start of experiment), middle (M, 21 days), and late (L, 42 days) stages of the experiment for microbiota composition analysis.
Detection of water physicochemical parameters
Water samples were collected from the overlying water of the aquaculture pond and outlet water of the reactor. Water samples (20 mL) were collected each time and detected within 2 h. Total nitrogen (TN) was measured using an LH-3BN total nitrogen analyzer (Lianhua Technology, China). COD was measured using a DR900 water quality analyzer (HACH, United States). Total phosphorus (TP), NH4-N, NO2-N, and nitrate (NO3-N) were determined via molybdenum antimony anti-spectrophotometry, Nessler reagent spectrophotometry, N-(1-naphthyl)-ethylenediamine spectrophotometry, and phenol disulfonic acid spectrophotometry, according to previously described methods, respectively (China Environment Publishing House, 2002).
Microbiota composition analysis using high-throughput sequencing of 16S rDNA
The water samples were filtered using 0.22-μm filter membranes to collect the microbiota. The biofilm microbiota were collected by placing the reactor packing samples into 50-mL centrifuge tubes containing 40-mL sterile ultrapure water, vibrating them at a 45-kHz ultrasonic frequency for 30 min, and then filtering the sterile ultrapure water using 0.22-μm filter membranes. The water and biofilm filter membranes were stored at −80°C. Water and biofilm microbial DNA were extracted using a DNeasy PowerSoil Pro kit (QIAGEN, Germany). DNA purity and concentration were determined using a Nanodrop 2000 spectrophotometer, and the DNA was diluted to 10 ng/μL for polymerase chain reaction (PCR) amplification. The V4-V5 hypervariable region was amplified in duplicate using the universal primer pair 515F and 909R with a 12-nucleotide sample-specific barcode included at the 5′-end of the 515F sequence to distinguish samples, as previously described (Xiang et al., 2018). PCR products were electrophoresed on a 1.2% agarose gel and purified using a SanPrep DNA gel recovery kit (Sangon Biotech, Shanghai, China). After all the purified DNA was mixed in equal amounts, Illumina HiSeq was used for pear-end sequencing according to a previously reported method. Raw reads were subjected to quality control and merged using QIIME 1.9.0 (Caporaso et al., 2010) and FLASH 1.2.8 (Magoc and Salzberg, 2011). Sequences with ≥97% similarity were clustered into operational taxonomic units (OTUs) using UPARSE (Edgar, 2013). The taxonomy of each OTU was assigned using the RDP classifier (Wang et al., 2007) according to the greengenes v13_8_99 reference files (DeSantis et al., 2006). OTU number, Chao1, Shannon, and Simpson indices were calculated using QIIME 1.9.0 to measure the α-diversity of microbiota.
Data analysis
Data are presented as mean ± standard deviation. The Kruskal–Wallis rank sum test combined with Dunn post hoc test was conducted using with the FSA package in R 4.2.0. Boxplots were drawn using the ggpubr package in R 4.2.0. Principal component analysis (PCA) was conducted using the STAMP software (Parks et al., 2014). A heatmap was drawn using the pheatmap package in R 4.2.0. Redundancy analysis (RDA) was conducted in R 4.2.0, using the vegan and ade4 packages. Correlations between biochemical parameters and microbes were assessed in R using the psych, reshape2, and corrplot packages. Linear discriminant analysis effect size (LEfSe) was conducted using the Galaxy platform (http://huttenhower.sph.harvard.edu/galaxy). Co-occurrence network analysis was conducted using the R igraph, psych, and Hmisc packages and graphed using the Gephi 0.9.2 software (Bastian et al., 2009). p < 0.05 indicated statistical significance.
Results
Changes in water physicochemical parameters during the experiment
During the first week of the experiment, the nitrate concentrations in the pond and reactor water showed upward trends and then gradually decreased. Twenty-eight days after the start of the experiment, it was stable and maintained at a concentration <2 mg/L until the end of the experiment (Figure 2A). At the beginning of the experiment, no significant difference was observed in the concentration of NO3-N in the water samples of each group. As the experiment continued until day 14, the concentration of NO3-N in the reactor water of groups B and C became significantly lower than that in the pond water. When the experiment continued until day 21, the concentrations of NO3-N in all reactors were significantly lower than that in the pond water. In addition, no significant difference was observed between the concentration of NO3-N in the reactor water of group A and that in the pond water on day 35. The NO3-N concentrations in other reactors were significantly lower than that in the pond water, and the trend lasted until the end of the experiment (Figure 2A). During the experiment, the NO2-N and NH4-N concentrations in the water sharply fluctuated, although the concentrations in the reactor water were significantly lower than those in the pond water at multiple time points (Figures 2B, C). The TN concentrations in the pond and reactors showed gradual downward trends during the experiment, and from day 21 until the end of the experiment, the TN concentrations in the group C reactors was significantly lower than that in the pond. The TN concentrations in the water samples of the group B reactors were also significantly lower than that in the pond water from day 28 until the end of the experiment (Figure 2D). These results indicated that the pond water itself had a certain self-purification ability to remove nitrogen, and the TN removal efficiency in the AW was accelerated by the reactor treatment. Adding Bacillus to the reactors improved their TN removal efficiency. However, the TP concentration in the reactor water sharply increased from approximately 1.5 mg/L to approximately 3.0 mg/L during the first 14 days and then showed a fluctuating downward trend. However, the TP concentration in the pond water sharply increased from day 14 and reached a stable state on day 28 of the experiment, with the concentration maintained at approximately 2.8 mg/L (Figure 2E). The COD in pond water showed a slight upward trend during the first 14 days and then showed a gradual downward trend. The COD in the reactor water fluctuated during the first 14 days and then showed a gradual downward trend. Moreover, from day 21, the COD in the reactor water of group C was significantly lower than that in the pond water, which lasted until the end of the experiment (Figure 2F). These results indicated that adding B. licheniformis to the reactor reduced the water COD.
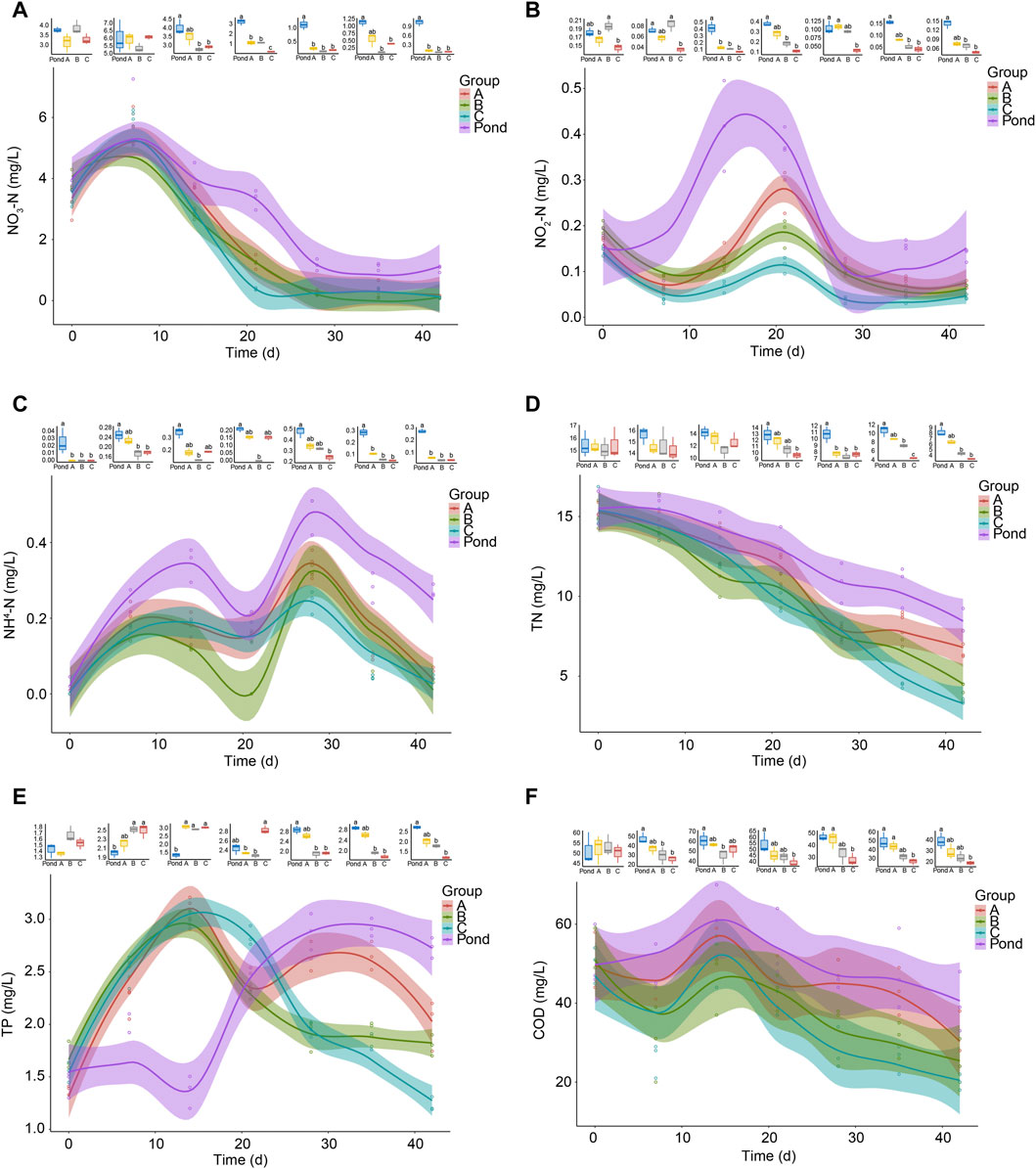
FIGURE 2. Changes in water quality of fish pond and reactor A, B, and C outlets. Group A is a natural biofilm group, and its biofilm was formed on the surface of the packing after the wastewater from the aquaculture pond continuously entered the reactors. Groups B and C are enhanced biofilm groups. In addition to providing aquaculture wastewater as in the group A reactors, 2.5 g of B. subtillis and B. licheniformis preparations was added to the two groups of devices every 3 days. The shaded areas around the curves represent the 95% confidence intervals. TN, total nitrogen; TP, total phosphorus; COD, chemical oxygen demand. Different letters above the boxes indicate that there were significant differences between the data. (A) NO3-N; (B) NO2-N; (C) NH4-N; (D) TN; (E) TP; and (F) COD. The boxplots above each part show the difference of indicators at each sampling time in fish pond, and reactor A, B, and C outlets. Distinct letters above the boxes indicate significant differences (p < 0.05).
Microbiota composition changes during experiment
Adding Bacillus to the reactor significantly increased the OTU number of the microbial community in the reactor water at the end of the experiment (Kruskal–Wallis rank sum test, p < 0.05; Figure 3A) and significantly increased the Chao1 index in the middle of the experiment (day 21; Kruskal–Wallis rank sum test, p < 0.05; Figure 3B). However, the Shannon and Simpson indices were not significantly affected (Kruskal–Wallis rank sum test, p > 0.05; Figures 3C, D). The addition of B. subtilis to the reactors significantly reduced OTU number and Chao1 index of the biofilm microbiota at the end of the experiment, as well as the Shannon and Simpson indices (Kruskal–Wallis rank sum test, p < 0.05), whereas the addition of B. licheniformis to the reactors did not affect the α-Diversity indices of the biofilm microbiota (Kruskal–Wallis rank sum test, p > 0.05; Figures 3A–D).
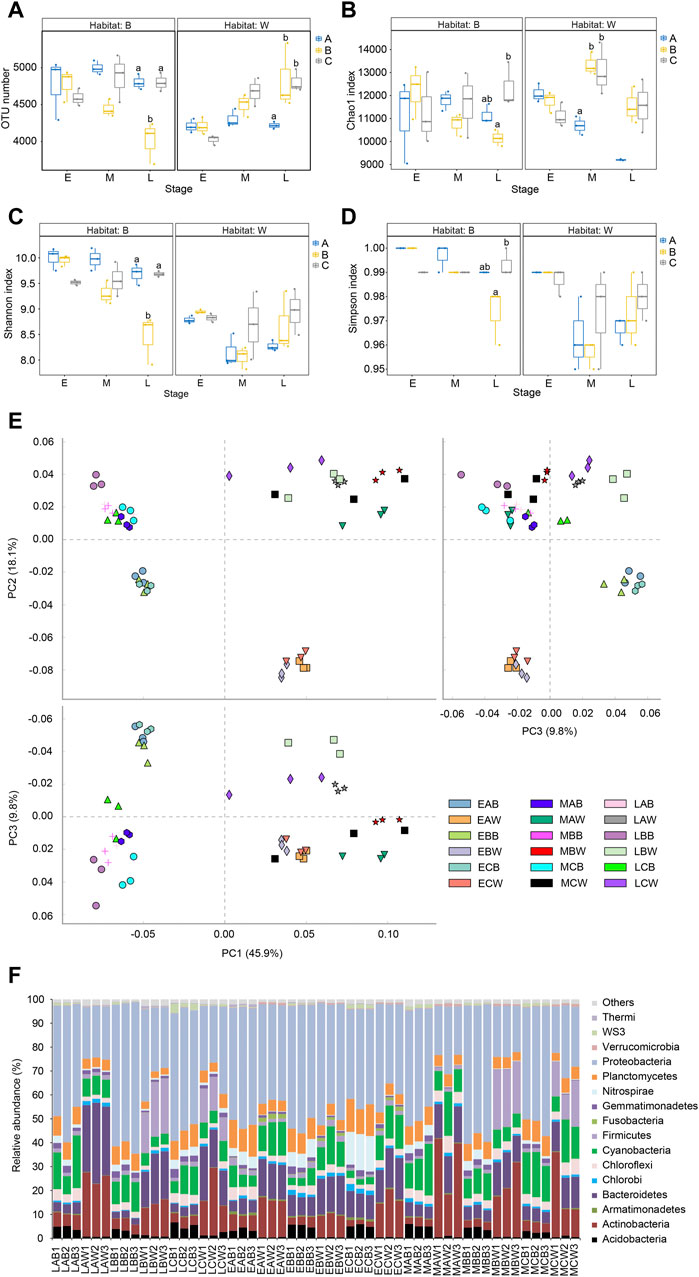
FIGURE 3. Alpha diversity indices and dominant phyla of water and biofilm of reactor packing microbiota. Group A is a natural biofilm group, and its biofilm was formed on the surface of the packing after the wastewater from the aquaculture pond continuously entered the reactors. Groups B and C are enhanced biofilm groups. In addition to providing aquaculture wastewater as in the group A reactors, 2.5 g of B. subtillis and B. licheniformis preparations was added to the two groups of devices every 3 days. Water (W) and biofilm of reactor packing B samples were collected at the early (E, 0 days after the start of experiment), middle (M, 21 days), and late (L, 42 days) stages of the experiment for microbiota composition analysis. (A) OTU number; (B) Chao index; (C) Shannon index; (D) Simpson index; (E) principal component analysis profile; (F) Composition of dominant phyla. Distinct letters above the boxes indicate significant differences (p < 0.05).
The PCA results showed that the samples were first divided into two groups according to water and biofilm, and the water samples were distributed in the first and fourth quadrants of the PCA profile drawn based on the first two axes, whereas the biofilm samples were distributed in the second and third quadrants (PERMANOVA, F = 43.684, p = 0.005). The samples were then grouped into different subgroups according to the sampling time. The samples collected at the beginning of the experiment were remarkably different from those collected at the middle and end stages of the experiment. The samples collected at the middle and end stages of the experiment were also significantly different (PERMANOVA, F = 20.108, p = 0.005 for the water microbiota; F = 14.692, p = 0.005 for the biofilm microbiota; Figure 3E).
Excluding the few sequences could not be determined at the phylum level, a total of 66 phyla (including 3 Archaea and 63 bacterial phyla) were detected, with Acidobacteria, Actinobacteria, Armatimonadetes, Bacteroidetes, Chlorobi, Chloroflexi, Cyanobacteria, Firmicutes, Fusobacteria, Gemmatimonadetes, Nitrospirae, Planctomycetes, Proteobacteria, Verrucomicrobia, WS3, and Thermi being the dominant microbiota (their relative abundance were more than 1% in at least one sample; Figure 3F). Adding both the B. subtillis and B. licheniformis preparations significantly increased the relative abundance of Chlorobi in the biofilm microbiota at the beginning of the experiment (Kruskal–Wallis rank sum test, p < 0.05; Supplementary Figure S1A) and significantly reduced the relative abundance of Thermi (Kruskal–Wallis rank sum test, p < 0.05; Supplementary Figure S1D); adding the B. licheniformis preparation significantly reduced the relative abundance of Firmicutes and Chloroflexi in the biofilm microbiota at the beginning of the experiment and significantly increased the relative abundance of Nitrospirae (Kruskal–Wallis rank sum test, p < 0.05; Supplementary Figure S1). Moreover, adding both the B. subtillis and B. licheniformis preparations significantly increased the relative abundance of Firmicutes in the water microbiota of the reactor at the middle and end stages of the experiment (Kruskal–Wallis rank sum test, p < 0.05; Supplementary Figure S1B).
Ninety of the 1,567 genera were detected as dominant genera (their relative abundance were more than 0.1% in at least one sample; Figure 4), which accounted for 76.31 ± 3.85% of the sequences. The samples were completely divided into two branches according to the microbial habitat: water and biofilm (Figure 4). The water samples were divided into three sub-branches according to the experimental stage (Figure 4). Although the biofilm samples collected at the early stage were completely separated from those collected at the middle and end stages, the samples collected at the middle and end stages were not separated from each other (Figure 4). At the early stage, the water samples were completely divided into three sub-branches according to the treatments. However, at the middle and end stages, only the water samples from group A were completely separated from those from the other two groups. The biofilm samples collected from the different stages were not completely divided according to treatment (Figure 4). Moreover, except for Novispirillum, other dominant genera differed significantly in their relative abundances between habitats, treatment groups, and experimental stages (Figure 4). However, these differences were primarily due to the habitat and experimental stage, and the addition of Bacillus had little impact on microbiota composition (Figure 4 and Supplementary Figure S2). However, the addition of Bacillus significantly increased the relative abundance of Bacillus in the water and biofilm microbiota at the middle and end stages of the experiment (Figure 4; Supplementary Figure S2A). The effects of adding B. subtilis and B. licheniformis on the main dominant genera in the reactor were not consistent, although both B. subtilis and B. licheniformis significantly increased the relative abundance of Rheinheimera in the reactor water microbiota in the middle and end stages of the experiment and significantly increased the relative abundance of Clostridium in the reactor water microbiota in the early stage of the experiment. The addition of B. licheniformis significantly reduced the relative abundance of Clostridium in the biofilm microbiota, whereas the addition of B. subtilis did not cause the same effect. The addition of B. subtilis and B. licheniformis reduced the relative abundance of Cylindrospermopsis, Flavobacterium, Fluviicola, and Gemmatimonas in the reactor water microbiota at the end of the experiment, whereas the addition of B. subtilis significantly reduced the relative abundance of Cylindrospermopsis in the reactor water microbiota in the middle stage of the experiment. The addition of B. licheniformis alone significantly increased the relative abundance of Nitrospira in the biofilm microbiota at the early stage of the experiment and in the water microbiota at the end of the experiment. Adding both B. subtilis and B. licheniformis significantly increased the relative abundance of Hyphomicrobium in the reactor water microbiota at the later stage of the experiment and Methylibium in the reactor water microbiota at the middle stage of the experiment; however, adding B. licheniformis alone significantly increased the relative abundance of Methylibium in the reactor water microbiota at the later stage of the experiment. Moreover, the addition of B. licheniformis significantly reduced the relative abundance of Hyphomicrobium in the biofilm microbiota at the early stage of the experiment, whereas the addition of B. subtilis significantly increased the relative abundance of Methylibium in the biofilm microbiota at the early stage of the experiment. Furthermore, the addition of B. subtilis and B. licheniformis had opposite effects on the relative abundance of Rhodococcus in the reactor water microbiota at the end of the experiment (Kruskal–Wallis rank sum test, p < 0.05; Supplementary Figure S2).

FIGURE 4. Dominant genera of dominant phyla of water and biofilm of reactor packing microbiota. Group A is a natural biofilm group, and its biofilm was formed on the surface of the packing after the wastewater from the aquaculture pond continuously entered the reactors. Groups B and C are enhanced biofilm groups. In addition to providing aquaculture wastewater as in the group A reactors, 2.5 g of B. subtillis and B. licheniformis preparations was added to the two groups of devices every 3 days. Water (W) and biofilm of reactor packing B samples were collected at the early (E, 0 days after the start of experiment), middle (M, 21 days), and late (L, 42 days) stages of the experiment for microbiota composition analysis. *** p < 0.001.
The LEfSe results showed that the addition of B. subtilis significantly increased the relative abundances of Clostridium, Rhodobacter, Giesbergeria, Hydrogenophaga, and Azospira in the water microbiota at the beginning of the experiment, Hyphomicrobium, Ralstonis, Rheinheimera, Acinetobacter, and Rhodococcus at the middle of the experiment, and Bacillus at the later stage of the experiment, and significantly increased the relative abundances of Planktothricoides, Cetobacterium, Gemmatimonas, Phenylobacterium, Rhodobacter, Giesbergeria, and Azospira in the biofilm microbiota at the beginning of the experiment, and Arthronema at the middle of the experiment. Adding B. licheniformis significantly increased the relative abundances of Microcystis, Phenylobacterium, and Rubrivivax in the water microbiota at the beginning of the experiment, and Nitrospira, Planctomyces, and Methylibium at the middle of the experiment, and significantly increased the relative abundances of Nitrospira, Planctomyces, Novosphingobium, Hydrogenophage, and Rubrivivax in the biofilm microbiota at the beginning of the experiment, Bacillus at the middle of the experiment, and Synechococcus later in the experiment (Supplementary Figure S3).
To analyze the potential correlations among bacteria in the community, the symbiotic network of microbial genera in the water column and the biofilm community was analyzed. Co-occurrence network analysis based on the Spearman coefficients of dominant genera showed that the co-occurrence of the water microbiota was considerably stronger than that of the reactor packing microbiota. Furthermore, although Bacillus was continuously added to groups B and C once every 3 days, the co-occurrence of Bacillus and other microbial genera was not the strongest, particularly in the reactor packing microbiota (Figure 5). These results indicated that Bacillus did not interact with these dominant bacteria directly, but indirectly through other closely related dominant bacteria in the microbiota.
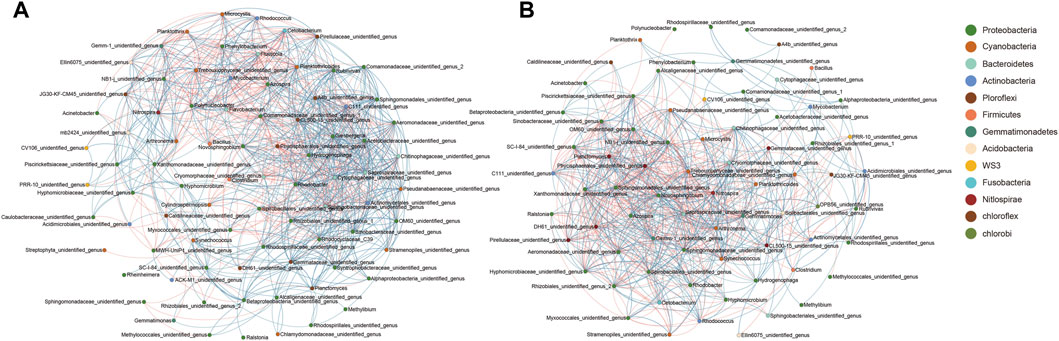
FIGURE 5. Co-occurrence networks showing the correlation between dominant genera in water (A) and reactor packing (B). The correlations with Spearman coefficient >0.6 and p < 0.05 are considered as significant. Orange and blue indicate significant negative and positive correlations, respectively.
The RDA results indicated that NO2-N and NH4-N levels were significantly correlated with the structure of the water microbiota in the reactors (Monte Carlo hypothesis test, p < 0.05; Figure 6A), and COD and TN, NO3-N, and NO2-N levels were significantly correlated with the structure of the biofilm microbiota on the surface of the reactor packing (Monte Carlo hypothesis test, p < 0.05; Figure 6B). Correlation analysis between the biochemical parameters and microbes showed that the relative abundance of Acinetobacter in the water microbiota of the reactor was significantly positively correlated with NH4-N and TP levels, whereas the relative abundance of Acinetobacter in the biofilm microbiota was significantly negatively correlated with TP. The relative abundance of Arthronema in both the water and biofilm was significantly negatively correlated with NO2-N levels, and the relative abundance of Arthronema in the water microbiota was significantly negatively correlated with TN and COD. The relative abundance of Azospira in both the water and biofilm microbiota was significantly positively correlated with NO2-N and NH4-N levels. The relative abundance of Bacillus in the water microbiota was significantly positively correlated with NO3-N, NO2-N, TN, and COD, whereas the relative abundance of Bacillus in the biofilm microbiota was significantly positively correlated with TN and COD. The relative abundance of Clostridium in the water microbiota was significantly positively correlated with NH4-N and TP levels, whereas the relative abundance of Clostridium in the biofilm microbiota was significantly positively correlated with TN. The relative abundance of Cylindrospermopsis in the water microbiota was significantly positively correlated with NO3-N levels and significantly negatively correlated with NH4-N levels, whereas the relative abundance of Cylindrospermopsis in the biofilm microbiota was significantly negatively correlated with NO2-N levels. The relative abundance of Giesbergeria in the water and biofilm microbiota was significantly positively correlated with NO2-N and NH4-N levels. The relative abundance of Hydrogenophaga in the water microbiota was significantly positively correlated with NO2-N and NH4-N levels. The relative abundance of Methylibium in the water microbiota was significantly negatively correlated with NO3-N, NO2-N, and TN levels and COD. The relative abundance of Nitrospira in the water microbiota was significantly negatively correlated with NO2-N levels, TN, and COD. The relative abundance of Planctomyces in the water microbiota was negatively correlated with TP, TN, and COD. The relative abundance of Ralstonia in the water microbiota was significantly positively correlated with NH4-N levels and TP, whereas the relative abundance of Ralstonia in the biofilm microbiota was significantly positively correlated with NO2-N levels and TN. The relative abundance of Rhodobacter in the water microbiota was significantly positively correlated with NH4-N levels (Figures 6C,D). Furthermore, the results indicated that the water physicochemical parameters of the reactors were more closely related to the water bacteria than biofilm bacteria. This result implied that the water microbiota were more susceptible to water physicochemical parameters than the biofilm microbiota.
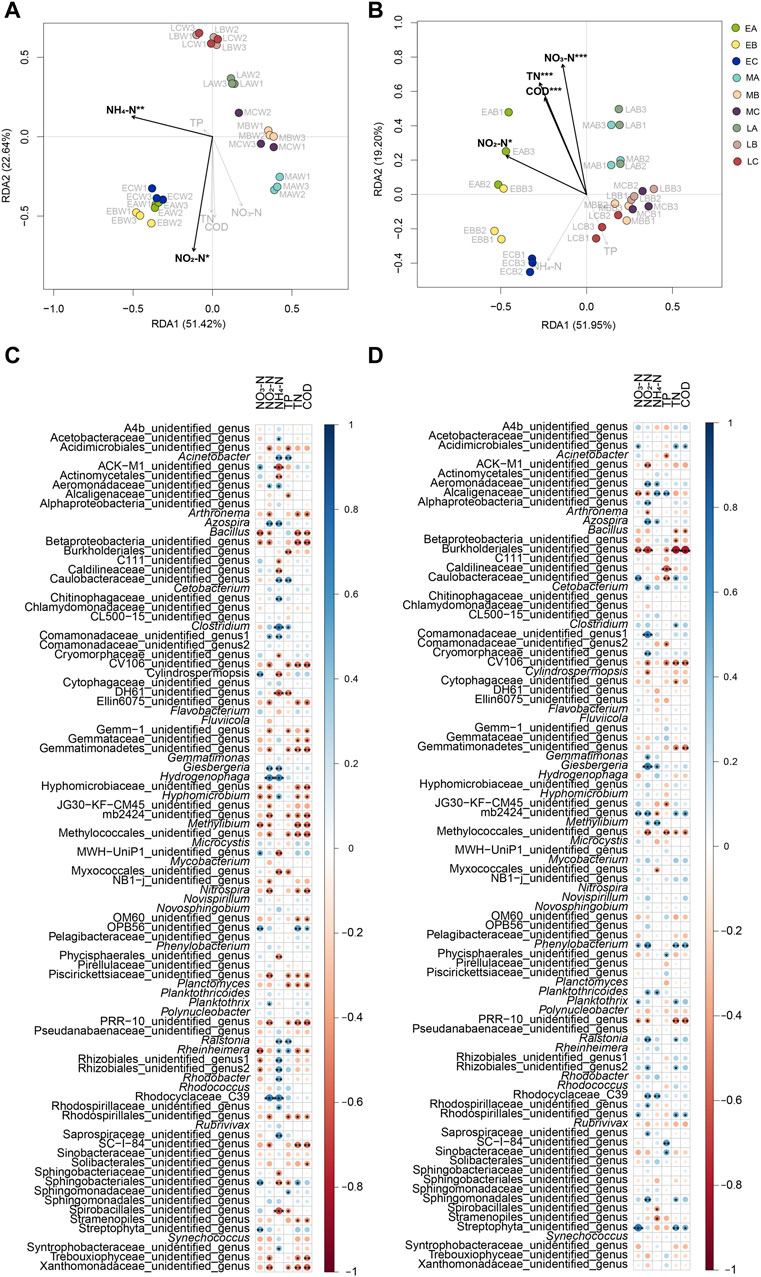
FIGURE 6. Correlation between water physicochemical parameters and both water and biofilm of reactor packing microbiota. (A) RDA profile showing correlation between water physicochemical parameters and water microbiota; (B) RDA profile showing correlation between water physicochemical parameters and biofilm microbiota; (C) Bubble chart showing correlation between water physicochemical parameters and water dominant genera; (D) Bubble chart showing correlation between water physicochemical parameters and biofilm dominant genera. Group A is a natural biofilm group, and its biofilm was formed on the surface of the packing after the wastewater from the aquaculture pond continuously entered the reactors. Groups B and C are enhanced biofilm groups. In addition to providing aquaculture wastewater as in the group A reactors, 2.5 g of B. subtillis and B. licheniformis preparations was added to the two groups of devices every 3 days. TN, total nitrogen; TP, total phosphorus; COD, chemical oxygen demand. Water (W) and biofilm of reactor packing B samples were collected at the early (E, 0 days after the start of experiment), middle (M, 21 days), and late (L, 42 days) stages of the experiment for microbiota composition analysis. * p < 0.05; ** p < 0.01; *** p < 0.001.
Discussion
Bacilli are Gram-positive aerobic or facultative anaerobic bacteria (Nayak, 2020). Because they can form spores that have excellent adaptability to different environments and are convenient for production and storage, they are typically used to develop microbial agents and repair environmental pollution problems (Chen and Hu, 2011). Bacillus have been widely used as environmental probiotics (Moriarty, 1998) in aquaculture due to their production of antimicrobial substances, enzymes provide and their ability to colonize the digestive tract and to contribute to nutrition of the host (Soltani et al., 2019). Han et al. (2021) used free and immobilized Bacillus cereus MRR2 to treat eutrophic water and found that the removal rates of phosphate, magnesium, and ammonium ions reached 90.1%, 95.6%, and 95.7% within 15 days, respectively. Lu et al. (2012) studied the repair effect of a Bacillus-matrix-plant integrated system on eutrophic water and found that the removal rates of Bacillus (1‰), limestone, Spartina alterniflora, and Iris tectorum on SRP were as high as 80.73%, and those of Bacillus (1.5‰), ceramsite, Aquilaria sinensis, and Iris tectorum on NH4-N and COD were 63.63% and 63.45%, respectively. Wang and Zhao (2011) used Bacillus coagulans YX-6 to purify shrimp culture wastewater and confirmed that B. coagulans YX-6 significantly improved the quality of the shrimp culture wastewater, increased the DO content, reduced the NH4-N content from 1.7 mg/L to 0.35 mg/L, reduced the NO2-N content from 0.64 mg/L to 0.05 mg/L, and reduced the average mortality of the shrimp. In this study, B. subtilis and B. licheniformis were added to grass carp culture wastewater purification reactors to increase the concentration of free-state Bacillus in the reactors, which significantly improved the removing efficiency of the TN and significantly reduced the COD of the wastewater.
Proteobacteria, Actinobacteria, and Bacteroidetes are the dominant bacteria in grass carp culture water (Zhang, 2014). In this study, by adding B. subtilis and B. licheniformis to a grass carp culture wastewater purification device, the relative abundance of Firmicutes in the middle and later stages of the experiment significantly increased. Many Firmicutes bacteria have a strong ability to remove nitrogen substances from water, and among them, Bacillus bacteria can promote the transformation of insoluble phosphorus into soluble phosphorus in water (Han et al., 2022). Jiang et al. (2021) also found that adding Bacillus to grass carp culture water in the early stage increased the TP in water. Therefore, this may also be the reason for the increase in TP in the water in this study. However, the addition of Bacillus did not increase the relative abundance of Firmicutes in the biofilm microbiota.
Most bacteria that had significant effects on the reactor water and biofilm microbiota after adding Bacillus were involved in the nitrogen cycle and organic matter metabolism. For instance, many Novosphingobium species exhibit good denitrification performance and are denitrifying bacteria (Zhou et al., 2016). Liu et al. (2021) reported that Novosphingobium was the dominant bacterial genus in a pig wastewater treatment plant, suggesting that the bacteria may have a strong wastewater treatment capacity. Bacillus and Acinetobacter also have a strong effect on the remediation of wastewater (Wei et al., 2016). Rhodococcus bacteria also contain nitrilases (Hoyle et al., 1998), and Rhodococcus can simultaneously perform heterotrophic nitrification and aerobic denitrification (Chen et al., 2012).
Biofilms are composed of a variety of microorganisms; therefore, there are complex interactions among microorganisms in biofilm bacterial communities, such as mutualism, symbiosis, parasitism, antagonism, and predation (Wen et al., 2021). Interactions between microorganisms can directly change the formation of biofilms (Xiao et al., 2021). Souza et al. (2020) promoted the growth of Streptococcus biofilms through cooperative co-culture between Candida albicans and Streptococcus. Synechococcus belongs to the phylum Cyanobacteria and is a primary producer in water. Synechococcus bacteria are widely distributed in marine and freshwater environments and can use dissolved organic matter in water for self-growth (Sun et al., 2005). Arthronema bacteria also belong to the phylum Cyanobacteria, which efficiently utilize carbon sources (Maheshwari et al., 2020); thus, they also have the ability to degrade organic matter in water. Wu et al. (2014) reported that Rubrivivax had good application prospects for the recycling of biomass resources and wastewater treatment. Hydrogenophaga exhibits strong degradation ability for polycyclic aromatic hydrocarbons (PAHs) and has an anaerobic denitrification function of reducing the concentration of nitrite in water (Meng et al., 2017). Rhodobacter has the ability to efficiently reduce the heavy metal chromium in water and reduce its toxicity (Rajyalaxmi et al., 2019). Planctomyces are highly advantageous among the microbiota of nitrogen and phosphorus removal systems (Wang et al., 2006), as they may have high nitrogen and phosphorus removal ability. The addition of Bacillus was found to significantly change the relative abundance of these bacteria. Simultaneously, the change in the relative abundance of these bacteria probably enhanced the efficiency of adding Bacillus to promote the removal of TN and reduction of COD in the AW treatment device, although the specific process and internal mechanism need to be further studied.
Uncultured members of the genus Nitrospira are the most diverse and abundant known nitrite-oxidizing bacteria in municipal wastewater treatment plants (Gruber-Dorninger et al., 2014; Cao et al., 2017) and soil (Hu et al., 2021). Moreover, recent findings have identified broader metabolic activity of Nitrospira (Koch et al., 2019), which were considered to be canonical nitrite-oxidizing bacteria with nitration capability (Mehrani et al., 2020). Although we found that the relative abundance of Nitrospira in the water microbiota was significantly negatively correlated with NO2-N levels, TN, and COD, the co-occurrence of Bacillus and Nitrospira did not provide similar results. These results implied that Bacillus may indirectly induce the participation of Nitrospira in nitrogen metabolism by regulating the entire microbiota.
The LEfSe method is widely used for the comparative analysis of microbiota compositions in various habitats and for the screening of significantly different microorganisms (Xiang et al., 2018; Li et al., 2019b; Li et al., 2019c; Mao et al., 2019; Liu et al., 2022). However, our results showed that this method could not simultaneously compare the differences between two or more treatment groups and the control. For instance, although the addition of both species of Bacillus significantly increased the relative abundance of Bacillus in the reactor water microbiota in the middle and later stages of the experiment (Supplementary Figure S2A), the LEfSe results only showed that the addition of B. subtilis significantly increased the relative abundance of Bacillus in the reactor water microbiota at the later stages of the experiment (Supplementary Figure S3A). These results indicated that the LEfSe method was not suitable for screening significantly different bacteria in two or more treatment groups being compared to the control.
Conclusion
Adding Bacillus bacteria to the reactors improved the TN removal efficiency of the reactors and reduced the COD. The addition of Bacillus changed the microbiota composition in the water and packing surface biofilm in the reactors, significantly increasing Bacillus in the water and biofilm microbiota in the middle and later stages of the experiment. However, the effects of adding B. subtilis and B. licheniformis on the main dominant genera in the reactor were not consistent.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA910292.
Author contributions
Conceptualization, ZLi and JX; Data curation, GW and XC; Formal analysis, ZLu and YG; Funding acquisition, ZLi, JX and XC; Investigation, ZLi, YG, ZLu, YL and GW; Methodology, ZLu, JX and XC; Project administration, JX and XC; Resources, ZLi, JX and XC; Software, ZLu and YG; Supervision, GW; Validation, YL; Visualization, ZLi and YG; Roles/Writing - original draft, ZLi; Writing - review & editing, JX and XC.
Funding
The Guangdong Basic and Applied Basic Research Foundation (grant number 2021A1515011116), National Natural Science Foundation of China (grant number 31802348), and the Central Public-interest Scientific Institution Basal Research Fund, CAFS (No.2022XT0503).
Acknowledgments
The authors would like to thank Bio-Form (Guangdong, China) for presenting the Bacillus subtilis and Bacillus licheniformis preparations. They are also grateful to Guangdong Meilikang Bio-Science Ltd., China for his assistance in data analysis.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fenvs.2023.1128329/full#supplementary-material
References
Bastian, M., Heymann, S., and Jacomy, M. (2009). “Gephi: An open source software for exploring and manipulating networks,” in International AAAI Conference on Weblogs and Social Media, San Jose, California, May 17, 2009 - May 20, 2009.
Cao, Y., van Loosdrecht, M. C. M., and Daigger, G. T. (2017). Mainstream partial nitritation-anammox in municipal wastewater treatment: Status, bottlenecks, and further studies. Appl. Microbiol. Biotech. 101, 1365–1383. doi:10.1007/s00253-016-8058-7
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi:10.1038/nmeth.f.303
Chen, C. C., and Chen, S. N. (2001). Water quality management with Bacillus spp. in the high density culture of red-parrot fish Cichlasoma citrinellum. C. Synspilum. N. Am. J. Aquacult. 63, 66–73. doi:10.1577/1548-8454(2001)063<0066:WQMWBS>2.0.CO;2
Chen, P., Li, J., Li, Q. X., Wang, Y., Li, S., Ren, T., et al. (2012). Simultaneous heterotrophic nitrification and aerobic denitrification by bacterium Rhodococcus sp. CPZ24. Bioresour. Technol. 116, 266–270. doi:10.1016/j.biortech.2012.02.050
Chen, S. Z., and Hu, Y. Y. (2011). Use of Bacillus subtilis in purification of slightly-polluted water. Acta Sci. Circumstantiae 31, 1594–1601. (in Chinese). doi:10.13671/j.hjkxxb.2011.08.002
China Environment Publishing House (2002). “Editorial committee of the state environmental protection administration of China on water and wastewater monitoring and analysis methods,” in Water and wastewater monitoring and analysis methods. Fourth Edition (Beijing: China Environment Publishing House).
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi:10.1128/AEM.03006-05
Edgar, R. C. (2013). Uparse: Highly accurate OTU sequences from microbial amplicon reads. Nat. Methods 10, 996–998. doi:10.1038/nmeth.2604
Edwards, P. (2015). Aquaculture environment interactions: Past, present and likely future trends. Aquaculture 447, 2–14. doi:10.1016/j.aquaculture.2015.02.001
Gruber-Dorninger, C., Pester, M., Kitzinger, K., Savio, D. F., Loy, A., Rattei, T., et al. (2014). Functionally relevant diversity of closely related Nitrospira in activated sludge. ISME J. 9, 643–655. doi:10.1038/ismej.2014.156
Han, Z., Guo, N., Yan, H., Xu, Y., Wang, J., Zhao, Y., et al. (2021). Recovery of phosphate, magnesium and ammonium from eutrophic water by struvite biomineralization through free and immobilized Bacillus cereus MRR2. J. Clean. Prod. 320, 128796. doi:10.1016/j.jclepro.2021.128796
Han, Z., Tang, C., Shao, L., Chen, S., Zhang, S., and He, P. (2022). The effect of multi-trophic freshwater ecosystem construction on microorganisms. J. Shanghai Ocean Univ. 2022. in press(in Chinese).
Hoyle, A. J., Bunch, A. W., and Knowles, C. J. (1998). The nitrilases of Rhodococcus rhodochrous MCIMB 11216. Enzyme Microb. Tech. 23, 475–482. doi:10.1016/s0141-0229(98)00076-3
Hu, J., Zhao, Y., Yao, X., Wang, J., Zheng, P., Xi, C., et al. (2021). Dominance of comammox Nitrospira in soil nitrification. Sci. Total Environ. 780, 146558. doi:10.1016/j.scitotenv.2021.146558
Ji, R., Lv, X., and Li, X. (2007). Performance of artificial medium for removing nutrients in eutrophic water. J. Lake Sci. 19, 39–45. doi:10.18307/2007.0106
Jiang, M., Zhang, W., Luo, L., Tang, X., Li, Y., Yu, X., et al. (2021). Effects of Bacillus methylotrophicus WM-1 on water quality dynamics and cultural fish in grass carp culture system. Aquac. Res. 52, 3371–3380. doi:10.1111/are.15182
Koch, H., van Kessel, M. A. H. J., and Lücker, S. (2019). Complete nitrification: Insights into the ecophysiology of comammox Nitrospira. Appl. Microbiol. Biotechnol. 103, 177–189. doi:10.1007/s00253-018-9486-3
Li, F., Feng, J., Zhou, X., Xu, C., Jijakli, M. H., Zhang, W., et al. (2019a). Impact of rice-fish/shrimp co-culture on the N2O emission and NH3 volatilization in intensive aquaculture ponds. Sci. Total Environ. 655, 284–291. doi:10.1016/j.scitotenv.2018.10.440
Li, H., Dong, X., Guo, G., Yang, X., Lin, Y., Cao, Y., et al. (2021). Effects of a strain Bacillus subtilis isolated from a culture pond on aquaculture water quality. Chin. J. Fish. 34, 73–78. (in Chinese).
Li, Z. F., Wang, G. J., Yu, E. M., Zhang, K., Yu, D. G., Gong, W. B., et al. (2019d). Artificial substrata increase pond farming density of grass carp (Ctenopharyngodon idella) by increasing the bacteria that participate in nitrogen and phosphorus cycles in pond water. Peer J. 7, e7906. doi:10.7717/peerj.7906
Li, Z. F., Wang, G. J., Zhang, K., Gong, W. B., Yu, E. M., Tian, J. J., et al. (2019c). Epizootic ulcerative syndrome causes cutaneous dysbacteriosis in hybrid snakehead (Channa maculata ♀ × Channa argus ♂). Peer J. 7, e6674. doi:10.7717/peerj.6674
Li, Z. F., Xie, J., Yu, E. M., Wang, G. J., Zhang, X. K., Yu, D. G., et al. (2019). Diet influences the accumulation of short-chain fatty acids associated with the gut microbiota in the grass carp (Ctenopharyngodon idellus) hindgut. Appl. Ecol. Env. Res. 17, 13435–13453. doi:10.15666/aeer/1706_1343513453
Liu, J., Yu, S., Cong, D., Yue, J., Yang, C., Wang, X., et al. (2021). Optimization of a novel single air-lift sequencing bioreactor for raw piggery wastewater treatment: Nutrients removal and microbial community structure analysis. Bioresour. Technol. 321, 124431. doi:10.1016/j.biortech.2020.124431
Liu, Q., Lai, Z., Wang, C., Ni, J., and Gao, Y. (2022). Seasonal variation significantly affected bacterioplankton and eukaryoplankton community composition in Xijiang River, China. Environ. Monit. Assess. 194, 55. doi:10.1007/s10661-021-09712-9
Lu, X. Z., Hao, J. C., Liu, H. F., and Tian, X. P. (2012). Study on the rehabilitation of eutrophic water by Bacillus–matrix–plant integrated system. Adv. Mat. Res. 599, 378–382. doi:10.4028/www.scientific.net/amr.599.378
Magoc, T., and Salzberg, S. L. (2011). Flash: Fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi:10.1093/bioinformatics/btr507
Maheshwari, N., Krishna, P. K., Thakur, I. S., and Srivastava, S. (2020). Biological fixation of carbon dioxide and biodiesel production using microalgae isolated from sewage waste water. Environ. Sci. Pollut. R. 27, 27319–27329. doi:10.1007/s11356-019-05928-y
Mao, L. T., Huang, J., Chen, Z. G., Ma, X. L., and Liu, H. R. (2019). Norfloxacin resistant bacterial compositions in sediments of Chinese subtropical fish ponds. Appl. Ecol. Env. Res. 17, 1039–1048. doi:10.15666/aeer/1701_10391048
Mehrani, M. J., Sobotka, D., Kowal, P., Ciesielski, S., and Makinia, J. (2020). The occurrence and role of Nitrospira in nitrogen removal systems. Bioresour. Technol. 303, 122936. doi:10.1016/j.biortech.2020.122936
Meng, J., Li, H., Tang, K., Song, K., and Feng, F. (2017). Analysis on naphthalene degradation characteristics of two Hydrogenophaga sp. strains. Environ. Prot. Chem. Ind. 37, 300–303. doi:10.3969/j.issn.1006-1878.2017.03.008
Moriarty, D. J. W. (1998). Control of luminous Vibrio species in penaeid aquaculture ponds. Aquaculture 164, 351–358. doi:10.1016/S0044-8486(98)00199-9
Nayak, S. K. (2020). Multifaceted applications of probiotic Bacillus species in aquaculture with special reference to Bacillus subtilis. Rev. Aquacult. 13, 862–906. doi:10.1111/raq.12503
Parks, D. H., Tyson, G. W., Hugenholtz, P., Beiko, R. G., and Notes, A. (2014). Stamp: Statistical analysis of taxonomic and functional profiles. Bioinformatics 30, 3123–3124. doi:10.1093/bioinformatics/btu494
Rajyalaxmi, K., Merugu, R., Girisham, S., and Reddy, S. M. (2019). Chromate reduction by purple non sulphur phototrophic bacterium Rhodobacter sp. GSKRLMBKU–o3 isolated from pond water. Proc. Natl. Acad. Sci. India, Sect. B Biol. Sci. 89, 259–265. doi:10.1007/s40011-017-0939-8
Soltani, M., Ghosh, K., Hoseinifar, S., Kumar, V., Lymbery, A. J., Roy, S., et al. (2019). Genus Bacillus, promising probiotics in aquaculture: Aquatic animal origin, bio-active components, bioremediation and efficacy in fish and shellfish. Rev. Fish. Sci. Aquac. 27, 331–379. doi:10.1080/23308249.2019.1597010
Souza, J. G. S., Bertolini, M., Thompson, A., Barão, V. A. R., and Dongari-Bagtzoglou, A. (2020). Biofilm interactions between Candida albicans and mitis group Streptococci in a titanium-mucosal interface model. Appl. Environ. Microb. 86, 029500–e3019. doi:10.1128/AEM.02950-19
Sun, S., Yue, H., and Xiao, T. (2005). Method and ecology function of marine Cyanobacteirum (Synechococcus). Sci. Tech. Eng. 5, 1628–1633. doi:10.3969/j.issn.1671-1815.2005.21.012
Tezzo, X., Bush, S. R., Oosterveer, P., and Belton, B. (2021). Food system perspective on fisheries and aquaculture development in Asia. Agr. Hum. values. 38, 73–90. doi:10.1007/s10460-020-10037-5
Wang, H., Zhou, Y., Dai, X., Chai, Y., Jiang, J., and Liu, S. (2006). Bacterial diversity study for the simultaneous nitrogen and phosphorus removal system (MDAT-IAT) by 16S rDNA cloning method. Acta Sci. Circumstantiae 26, 903–911. (in Chinese).
Wang, J., and Zhao, L. (2011). Study on the application of Bacillus coagulans to purify aquaculture water. Adv. Mat. Res. 343, 988–993. doi:10.4028/www.scientific.net/amr.343-344.988
Wang, M. M., Wei, J., Huang, R. M., Kong, L. W., Li, Z. Z., and Yang, B. (2021). Research progress of subsurface flow constructed wetland filler and its pollutant removal mechanism. J. Environ. Eng. Technol. 11, 769–776. doi:10.12153/j.issn.1674-991X.20200199
Wang, P., Ji, J., and Zhang, Y. (2020). Aquaculture extension system in China: Development, challenges, and prospects. Aquacult. Rep. 17, 100339. doi:10.1016/j.aqrep.2020.100339
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microb. 73, 5261–5267. doi:10.1128/AEM.00062-07
Wei, J., Gong, X., Li, X., Yan, Y., Li, Y., Zhao, J., et al. (2016). Research on the diversity of microbial community structure of tail water from sewage plants in different areas. J. Anhui Agri. Sci. 44, 23–25. (in Chinese). doi:10.13989/j.cnki.0517-6611.2016.11.008
Wen, J., Xiao, Y., Song, P., Sun, B., Muhammad, T., Ma, L., et al. (2021). Bacillus amyloliquefaciens application to prevent biofilms in reclaimed water microirrigation systems. Irrig. Drain. 70, 4–15. doi:10.1002/ird.2527
Wu, G., Gao, M., Tang, J., Lu, H., and Wu, Y. (2019). Progress of researches on nitrogen and phosphorus removal by periphytic biofilm from non-point source wastewater. J. Eco. Rural. Environ. 35, 817–825. doi:10.19741/j.issn.1673-4831.2019.0035
Wu, P., Li, J., Wang, Y., Du, C., Tong, Q., Liu, X., et al. (2014). Strengthening the growth of Rubrivivax gelatinosus in sewage purification through ferric ion regulated photophosphorylation and respiration. Water Sci. Technol. 70, 1969–1975. doi:10.2166/wst.2014.440
Wu, Y., Liu, J., Yang, L., Chen, H., Zhang, S., Zhao, H., et al. (2011). Allelopathic control of cyanobacterial blooms by periphyton biofilms. Environ. Microbiol. 13, 604–615. doi:10.1111/j.1462-2920.2010.02363.x
Xiang, J., He, T., Wang, P., Xie, M., Xiang, J., and Ni, J. (2018). Opportunistic pathogens are abundant in the gut of cultured giant spiny frog (Paa spinosa). Aquac. Res. 49, 2033–2041. doi:10.1111/are.13660
Xiao, Y., Liu, Y., Ma, C., Muhammad, T., Zhou, B., Zhou, Y., et al. (2021). Using electromagnetic fields to inhibit biofouling and scaling in biogas slurry drip irrigation emitters. J. Hazard. Mat. 401, 123265. doi:10.1016/j.jhazmat.2020.123265
Yi, Y., Li, Y., Rong, Y., and Li, K. (2020). Decontamination of aquaculture sewage by microorganisms. Hubei Agric. Sci. 49, 2509–2511. (in Chinese).
Zhang, H. (2011). “Development of new type of Bacillus probiotics and its application in eutrophic water,” (Guangzhou: Jinan University). Master’s thesis.
Zhang, X. (2014). “Effects of Bacillus subtilis SC02 and Pseudomonas stutzeri F1M on water quality in grass carp (Ctenopharyngodon idellus) culture and its mechanism,” (Hangzhou: Zhejiang University). Doctoral dissertation.
Zhou, S., Huang, T., Bai, S., and He, X. (2016). Isolation, identification, and nitrogen removal characteristics of oligotrophic aerobic denitrifiers. Chin. Environ. Sci. 36, 238–248. (in Chinese). doi:10.1007/s11771-009-0240-y
Keywords: Bacillus subtilis, Bacillus licheniformis, natural biofilm, aquaculture wastewater treatment, microbiota
Citation: Li Z, Gao Y, Lu Z, Xie J, Liu Y, Wang G and Cheng X (2023) Strengthening and microbial regulation mechanism of Bacillus on purification device for grass carp culture wastewater. Front. Environ. Sci. 11:1128329. doi: 10.3389/fenvs.2023.1128329
Received: 20 December 2022; Accepted: 13 January 2023;
Published: 25 January 2023.
Edited by:
Sherif Ismail, Shandong University, ChinaReviewed by:
Shiyang Zhang, Wuhan University of Technology, ChinaJunfeng Chen, Qufu Normal University, China
Copyright © 2023 Li, Gao, Lu, Xie, Liu, Wang and Cheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangju Cheng, chengxiangju@scut.edu.cn
 Zhifei Li
Zhifei Li Yan Gao2
Yan Gao2  Jun Xie
Jun Xie Guangjun Wang
Guangjun Wang